Merge pull request #3946 from RosettaCommons/vmullig/rosetta_thread_manager
Implement a Rosetta thread manager
In Rosetta, we have many different levels at which we might want to launch threads. For example, a job distributor could try to carry out jobs in parallel threads. A mover might parallelize its own work. Low-level functions, like interaction graph setup for the packer, gradient vector calculation for the minimizer, or scorefunction evaluation could also be parallelized over threads. So what happens when modules at many different levels all try to spawn threads? You could end up with a nasty thread explosion. Imagine, for example, that my job distributor launches 16 threads, each of which calls a mover that launches 16 threads, each of which calls another mover that launches 16 threads, each of which calls the packer, which launches 16 threads. You'd end up with 65,536 threads all contending for hardware resources.
Another consideration is that the master/slave relationship that tends to work well for MPI communication isn't ideally suited to threads, where you might just have 4 or 8 cores available, all of which should be _doing_ work rather than _managing_ work. So we want something that ensures that many different layers of Rosetta can ask for threads, without stepping on one another's toes, _and_ we want that thing not to hog resources to monitor everything.
In addition, thread-based parallelism, unlike process-based parallelism, works best with small, finely-grained parallel tasks that are all accessing similar regions of memory, which means that a certain amount of synchronization makes sense. Asynchronous job-level parallelism might make more sense with MPI calls than with threads -- I'm not sure. I want us to be able to experiment with parallelism on many different levels, but right now, we risk stepping on one another's toes if I'm parallelizing the packer and someone else is parallelizing a mover and someone else is writing a parallel job distributor.
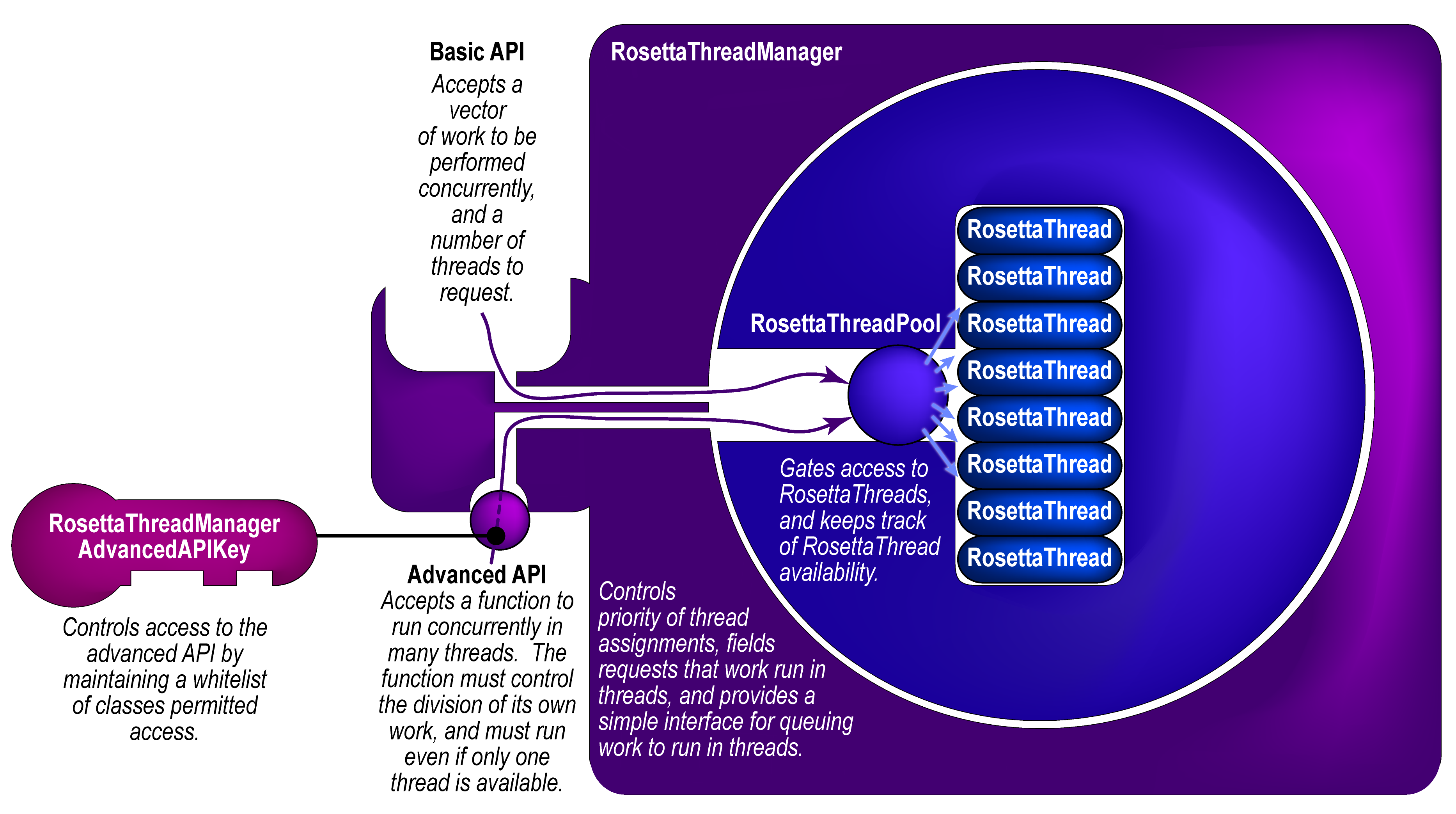
I'm proposing the `RosettaThreadManager` as a solution to this. Given N total threads that the user wants to run at any given time (assuming he or she has N cores on his or her system), the global `RosettaThreadManager` spins up and maintains a pool of N-1 threads. When a module wants to do something in parallel (either from the master thread or from a child thread), it bundles the function that it wants to run in threads with its arguments using `boost::bind`, then passes that function to the `RosettaThreadManager` with a request for M threads. The `RosettaThreadManager` launches that process _synchronously_ on available idle threads, including the calling thread, with a firm guarantee that 1 <= number of threads assigned to the function <= min(total threads, number requested). The function is responsible for carrying out its work, with its parallel siblings, in a threadsafe manner. At the end, the requesting thread blocks until all sibling functions terminate, the threads are released to idle status, and other work can run on them.
Based on community feedback, I have also added a basic API that accepts a vector of work to be done. The advanced API, which allows an arbitrary thread function to run concurrently, is now access-gated with a key class that has a private constructor, so that only whitelisted friend classes can use the advanced API. This forces developers _either_ to use the basic API, _or_ to justify to the community why their class should be allowed access to the advanced API.
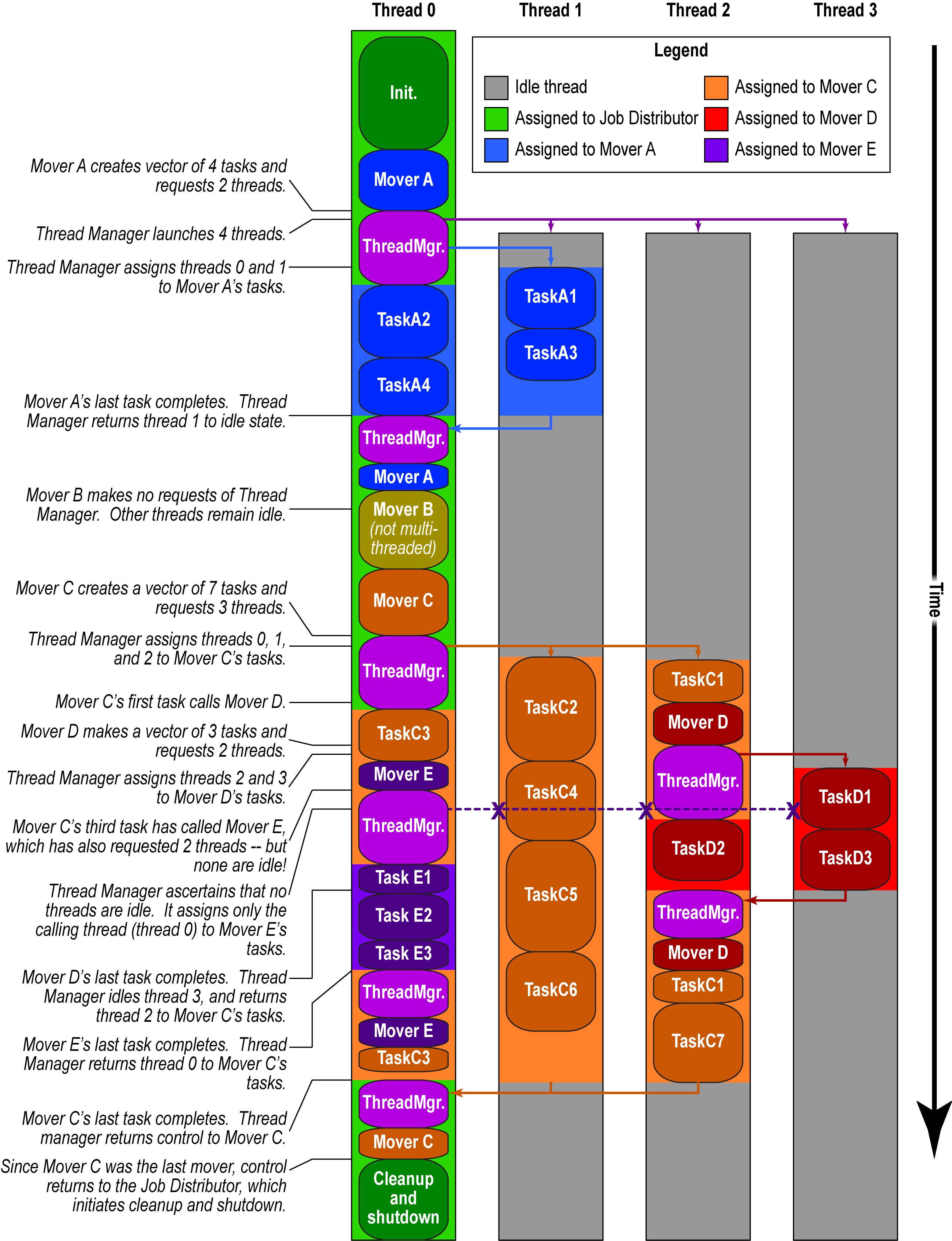
This PR implements the basic infrastructure for this, as well as an application that shows the effects of launching threads at three different levels. In the app, a master thread runs a level 1 function in N threads, and each level 1 function runs M level 2 functions, and each level 2 function runs P level 3 functions. The level 3 functions all work together to calculate a times table (though I could just as easily have only those level 3 functions that were launched by a single level 2 function coorperate on a piece of data). When all levels return, the master thread checks that the times table was calculated correctly, in parallel.
TODO:
- [x] Write `RosettaThreadAssignmentInfo` class.
- [x] Finish `RosettaThreadManager` class.
- ~~Wait for, and spin down, all running threads on `RosettaThreadPool` destruction. (Probably not necessary given the model of ensuring that functions are run in both the calling thread and assigned threads, and the calling thread waits for assigned threads to finish before returning, but could prevent fragility in the future if the model changes.)~~ --> I realized that this might cause the app to hang when an exception is thrown, instead of exiting properly... --> Never mind. I did get this to work sensibly.
- [x] Means of getting a thread's thread ID.
- [x] Test app.
- [x] Talk to Sergey about cxx11thread-mode integration tests.
- [x] Pull request #3957 adds support for cxx11thread-mode integration tests. That pull request must be merged before this one. (This one has that one merged into it.)
- [x] Make this into a unit/integration test.
- [x] Add information to the `RosettaThreadAssignmentInfo` class about the level from which a multithreading request comes, in case we want to make decisions about thread assignments based on that in the future.
- [x] Enums for app-level, job distributor-level, mover-level, filter-level, taskop-level, resselector-level, simplemetric-level, core compenent level.
- [x] Beauty.
- [ ] Add developer docs.
- [ ] Document what `{?}` means.
- [x] Switch from `boost::bind` and `boost::function` to `std::bind` and `std::function`.
- [x] Switch to `condition_variable` instead of using a wait in a loop.
Additional changes:
- [x] Switch enum to enum class (see Sergey's comments).
- [x] Switch to crash with nonzero exit code on thread termination failure.
For a future pull request:
- Parallelize the interaction graph set-up.
- Parallelize `GeneralizedKIC`.
- Switch the `MultithreadedJobDistributor` to use this infrastructure.
- Integration test for the above three things.